Making a main field model#
Here we demonstrate how to make a simple time-independent main field model by fitting spherical harmonics to ground observatory measurements.
The principles of spherical harmonic analysis have been shown in the previous pages, so here we will take a shortcut and use utilties from ChaosMagPy.
ChaosMagPy, Clemens Kloss, https://doi.org/10.5281/zenodo.3352398
We will use:design_gauss to generate the design matrix for the inversion
synth_values to run the forward model to get predictions for the magnetic field from our model
hvPlot is used to create some fancy visualisations but this library is more complex to use! so if you are newer to the Python ecosystem, you are better off using Matplotlib for your own work.
# Install pooch (not currently in VRE)
import sys
!{sys.executable} -m pip install pooch
Show code cell output
Requirement already satisfied: pooch in /opt/conda/lib/python3.9/site-packages (1.7.0)
Requirement already satisfied: packaging>=20.0 in /opt/conda/lib/python3.9/site-packages (from pooch) (21.3)
Requirement already satisfied: requests>=2.19.0 in /opt/conda/lib/python3.9/site-packages (from pooch) (2.27.1)
Requirement already satisfied: platformdirs>=2.5.0 in /opt/conda/lib/python3.9/site-packages (from pooch) (2.5.0)
Requirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /opt/conda/lib/python3.9/site-packages (from packaging>=20.0->pooch) (3.0.7)
Requirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.9/site-packages (from requests>=2.19.0->pooch) (2021.10.8)
Requirement already satisfied: idna<4,>=2.5 in /opt/conda/lib/python3.9/site-packages (from requests>=2.19.0->pooch) (3.3)
Requirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.9/site-packages (from requests>=2.19.0->pooch) (1.26.8)
Requirement already satisfied: charset-normalizer~=2.0.0 in /opt/conda/lib/python3.9/site-packages (from requests>=2.19.0->pooch) (2.0.12)
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv
[notice] A new release of pip is available: 23.0.1 -> 23.1.2
[notice] To update, run: pip install --upgrade pip
import pooch
from chaosmagpy.model_utils import design_gauss, synth_values, power_spectrum
from chaosmagpy.plot_utils import plot_power_spectrum
import numpy as np
import pandas as pd
import xarray as xr
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import hvplot.xarray
Loading some data#
We’ll load a pre-prepared dataset containing annual means derived from observatory data. We just select a single year from this dataset. In reality the main field varies on shorter time scales but these annual data points are useful enough for a low resolution model.
# Download annual means dataset
# https://geomag.bgs.ac.uk/data_service/data/annual_means.shtml
oamjumpsapplied = pooch.retrieve(
"https://raw.githubusercontent.com/MagneticEarth/IAGA_SummerSchool2019/master/data/external/oamjumpsapplied.dat",
known_hash="dee77ded79cb0d86e3366a550830cad9de741322212faf9a74f3d03cb6bdd872"
)
def load_data(year=2000.5):
df = pd.read_csv(oamjumpsapplied, delim_whitespace=True)
df = df.where(df["year"] == 2000.5).dropna()
df = df.reset_index(drop=True)
# Screen out some bad data points
df = df.where(df["Z"] != 88888).dropna()
_ds = df.to_xarray()
_ds["Latitude"] = 90 - _ds["Colat"]
_ds["Longitude"] = (_ds["Long"] % 360 + 540) % 360 - 180
_ds = _ds.drop("index")
return _ds
input_data = load_data(year=2000.5)
input_data
<xarray.Dataset>
Dimensions: (index: 164)
Dimensions without coordinates: index
Data variables: (12/14)
Code (index) object 'ALE' 'NAL' 'CCS' 'THL' ... 'VNA' 'SBA' 'VOS'
Colat (index) float64 7.5 11.08 12.28 12.52 ... 158.6 160.7 167.8 168.4
Long (index) float64 297.6 11.93 104.3 290.8 ... 351.8 166.8 106.9
alt(m) (index) float64 60.0 12.0 10.0 57.0 ... 0.0 40.0 10.0 3.5e+03
year (index) float64 2e+03 2e+03 2e+03 2e+03 ... 2e+03 2e+03 2e+03
D (index) float64 -67.51 0.332 14.76 -63.17 ... -12.91 155.3 -120.7
... ...
X (index) float64 1.319e+03 7.263e+03 ... -1.018e+04 -6.859e+03
Y (index) float64 -3.186e+03 42.0 746.0 ... 4.692e+03 -1.156e+04
Z (index) float64 5.555e+04 5.384e+04 ... -6.622e+04 -5.819e+04
F (index) float64 5.566e+04 5.433e+04 ... 6.716e+04 5.972e+04
Latitude (index) float64 82.5 78.92 77.72 77.48 ... -70.65 -77.85 -78.45
Longitude (index) float64 -62.35 11.93 104.3 -69.17 ... -8.25 166.8 106.9def plot_obs_measurements(ds=input_data):
return ds.hvplot.scatter(
x="Longitude", y="Latitude", c="Z",
coastline=True, projection=ccrs.PlateCarree(), global_extent=True,
clabel="Data: vertical (downwards) magnetic field (nT)"
)
plot_obs_measurements(input_data)
WARNING:param.Parameterized: Use method 'warning' via param namespace
WARNING:param.main: geo option cannot be used with kind='scatter' plot type. Geographic plots are only supported for following plot types: ['bivariate', 'contour', 'contourf', 'dataset', 'hexbin', 'image', 'labels', 'paths', 'points', 'points', 'polygons', 'quadmesh', 'rgb', 'vectorfield']
/opt/conda/lib/python3.9/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
Building a model#
Suppose we have data,
Note that:
A least-squares solution to
def build_model(ds=input_data, nmax=10):
"""Returns Gauss coefficients for a simple SH model"""
theta = ds["Colat"]
phi = ds["Long"]
radius = 6371.2 + ds["alt(m)"]/1e3
B_radius = -ds["Z"]
B_theta = -ds["X"]
B_phi = ds["Y"]
# Build the design matrix
G_radius, G_theta, G_phi = design_gauss(radius, theta, phi, nmax=nmax, source="internal")
# Perform the inversion
G = np.concatenate((G_radius, G_theta, G_phi))
d = np.concatenate((B_radius, B_theta, B_phi))
m = np.linalg.inv(G.T @ G) @ (G.T @ d)
return m
m = build_model(input_data, nmax=5)
m
array([-2.96482406e+04, -1.76621579e+03, 5.06863059e+03, -2.47450873e+03,
3.13168299e+03, -2.64941275e+03, 1.74507015e+03, -3.76171556e+02,
1.10537201e+03, -2.25018745e+03, -6.30954563e+01, 1.19694272e+03,
3.54883214e+02, 8.12814264e+02, -5.15153390e+02, 9.99590132e+02,
9.11464975e+02, 1.54019008e+02, 3.42255212e+02, -3.26457915e+02,
-3.49774227e+02, 2.34546359e+02, 7.81036147e+01, -3.73915172e+02,
-2.86846063e+02, 1.90610037e+02, -1.94545936e+02, 3.69767832e+02,
1.49286822e+02, -1.94868808e+02, -7.07565425e+01, -1.49668101e+02,
-2.26259266e+01, -5.93299060e+01, 1.62121406e+02])
Since we do not have many data, we would need to be more sophisticated with the inversion procedure (e.g. including regularisation) to get a model of higher resolution. For this reason, the model is truncated at spherical harmonic degree 6. You can explore increasing this limit.
How good is our model?#
Evaluating a model over the Earth and plotting contours…
def sample_model_on_grid(m, radius=6371.200):
"""Evaluate Gauss coefficients over a regular grid over Earth"""
theta = np.arange(1, 180, 1)
phi = np.arange(-180, 180, 1)
theta, phi = np.meshgrid(theta, phi)
B_r, B_t, B_p = synth_values(m, radius, theta, phi)
ds_grid = xr.Dataset(
data_vars={"B_NEC_model": (("y", "x", "NEC"), np.stack((-B_t, B_p, -B_r), axis=2))},
coords={"Latitude": (("y", "x"), 90-theta), "Longitude": (("y", "x"), phi), "NEC": np.array(["N", "E", "C"])}
)
return ds_grid
def plot_model_contours(m):
ds_grid = sample_model_on_grid(m)
# Generate contour plot of vertical component
return ds_grid.sel(NEC="C").hvplot.contourf(
x="Longitude", y="Latitude", c="B_NEC_model",
levels=30, coastline=True, projection=ccrs.PlateCarree(), global_extent=True,
clabel="Model: vertical (downwards) magnetic field (nT)"
)
plot_model_contours(m)
/opt/conda/lib/python3.9/site-packages/geoviews/operation/projection.py:79: ShapelyDeprecationWarning: Iteration over multi-part geometries is deprecated and will be removed in Shapely 2.0. Use the `geoms` property to access the constituent parts of a multi-part geometry.
polys = [g for g in geom if g.area > 1e-15]
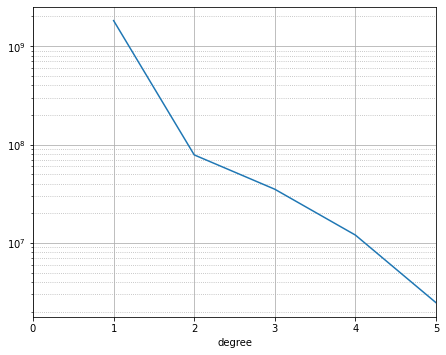
The power spectrum is a common way to assess and compare models, showing how much power is concentrated at each spherical harmonic degree:
plot_power_spectrum(power_spectrum(m));
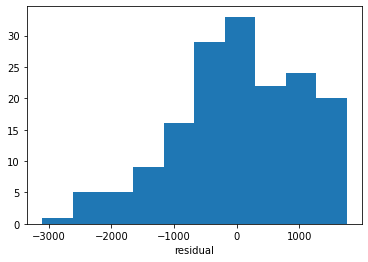
… and we can look at the residuals between the input data and the model:
def append_residuals(ds=input_data, m=m):
# Evaluate model at the data locations
B_r, B_t, B_p = synth_values(m, 6371.2 + ds["alt(m)"]/1e3, ds["Colat"], ds["Longitude"])
# Assign data-model residuals to the dataset
ds["res_N"] = ds["X"] - (-B_t)
ds["res_E"] = ds["Y"] - (B_p)
ds["res_C"] = ds["Z"] - (-B_r)
return ds
ds = append_residuals(input_data, m)
plt.hist(ds["res_C"])
plt.xlabel("residual");
ds.hvplot.scatter(
x="Longitude", y="Latitude", c="res_C",
coastline=True, projection=ccrs.PlateCarree(), global_extent=True,
clabel="Data-model residual: vertical (downwards) magnetic field (nT)"
)
WARNING:param.Parameterized: Use method 'warning' via param namespace
WARNING:param.main: geo option cannot be used with kind='scatter' plot type. Geographic plots are only supported for following plot types: ['bivariate', 'contour', 'contourf', 'dataset', 'hexbin', 'image', 'labels', 'paths', 'points', 'points', 'polygons', 'quadmesh', 'rgb', 'vectorfield']
Exercises#
Experiment with changing the SH degree trunction in the model
Experiment with using data just over Europe
Add random noise into the data to observe the effect
Use viresclient to access the monthly means dataset and produce a model using those instead
def select_europe(ds=input_data):
"""Subselect the dataset down to just cover Europe"""
colat_minmax = (20, 60)
lon_minmax = (-10, 40)
ds = ds.where(
(ds["Colat"] > colat_minmax[0]) & (ds["Colat"] < colat_minmax[1]) &
(ds["Long"] > lon_minmax[0]) & (ds["Long"] < lon_minmax[1]),
).dropna(dim="index")
return ds
